PrimeTime™ qPCR Primer Assays
Premixed primer pairs for analyzing gene expression using intercalating dyes.
PrimeTime qPCR Primer Assays provide a primer pair designed for real-time PCR using intercalating dyes, such as SYBR® Green (Thermo Fisher Scientific) or EvaGreen® (Biotium) dyes. Predesigned sequences for human, mouse, or rat are designed with advanced bioinformatic and thermodynamic sequence analytics and for easy selection. Custom assays may be created for any sequence from any species using the PrimerQuest™ Tool.
Ordering
- Advanced bioinformatics delivers the design you need with considerations for exon location and number of transcripts identified
- Predesigned sequences will achieve ≥90% efficiency or we will provide an alternative design free of charge
- Test target sequence amplification prior to using more expensive probe-based assays
- Receive primer sequences with all orders
PrimeTime qPCR Primer Assays in tubes
Available in standard size, premixed, and shipped dry.
|
1 Available for human, mouse, and rat targets (for assay configuration, choose intercalating dyes, primers only)
2 Choose design: qPCR–2 primers–intercalating dyes
PrimeTime qPCR Primer Assays in 96-well plates
Custom assays and predesigned qPCR sequences for human, mouse, and rat targets. Available in standard size, premixed, and shipped dry.
|
Product details
PrimeTime qPCR Primer Assays can be used with SYBR® Green (Thermo Fisher Scientific), EvaGreen® (Biotium), and other intercalating dyes, where no probe is needed. PrimeTime qPCR Primer Assays are available in standard size, premixed, normalized to 5 nmol per primer, and shipped dried down. Primer sequences are provided upon order.
PrimeTime qPCR Primer Assays
- The same primer pairs found in the PrimeTime qPCR 5' Nuclease Assays
- Forward and reverse primers, mixed and delivered in a single tube or plate well
- Compatible with SYBR® Green, EvaGreen®, and other intercalating dyes, where no probe is needed
- Available in tubes or 96-well plates
Predesigned sequences are designed using a proprietary algorithm. The bioinformatic calculations optimizes oligo melting temperature (Tm), base composition, oligo length, etc., and accounts for factors such as SNPs (based on current NCBI RefSeq releases), off-target amplification, splice variants, and secondary structures.
Guarantee: Predesigned sequences will achieve 90% efficiency or better, or we will provide an alternative design free of charge. For more information, please contact us.
Custom primer assays can also be designed for other species using the PrimerQuest™ Tool. This tool may be used to design oligos for endpoint PCR, qPCR, and Sanger sequencing. Use our preset design parameters for PCR and qPCR or customize parameters for your application. The PrimerQuest Tool is based on the Primer3 engine.
For commonly studied pathways in human, mouse, and rat species, we have suggested gene sets (see Resources section below) that can be used with our PrimeTime plate ordering system.
For help with assay design, contact us.
Primer sequences are provided
We provide primer sequences with each order to assist with best practices in research reporting.
- Boosts publication credibility—you can publish your research according to MIQE guidelines
- Helps identify and avoid SNPs
- Facilitates design of multiplex experiments
- Assists with data interpretation and troubleshooting
- Allows you to validate your transcripts
Product data
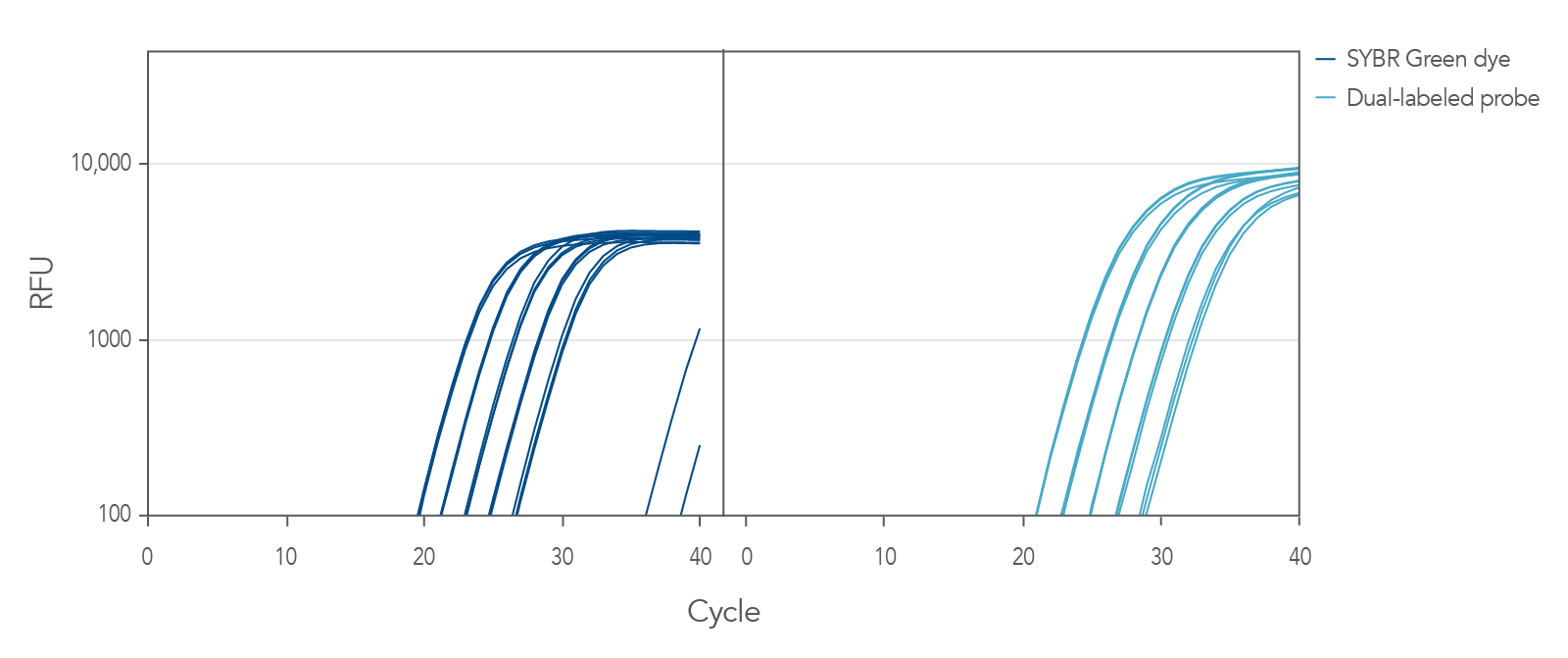
Figure 1. PrimeTime qPCR Primer Assays yield the same high efficiency whether used with intercalating dyes or probes. Amplification of 5 sequential 4-fold dilutions of cDNA using PrimeTime qPCR Primer Assays (with SYBR® Green dye [Bio-Rad SsoAdvanced Universal Probes Supermix]) or the PrimeTime qPCR 5′ Nuclease Assay (with dual-labeled probe [IDT PrimeTime Gene Expression Master Mix]) targeting human v-abl Abelson murine leukemia viral oncogene homolog 1 (ABL1, NM_005157.5).
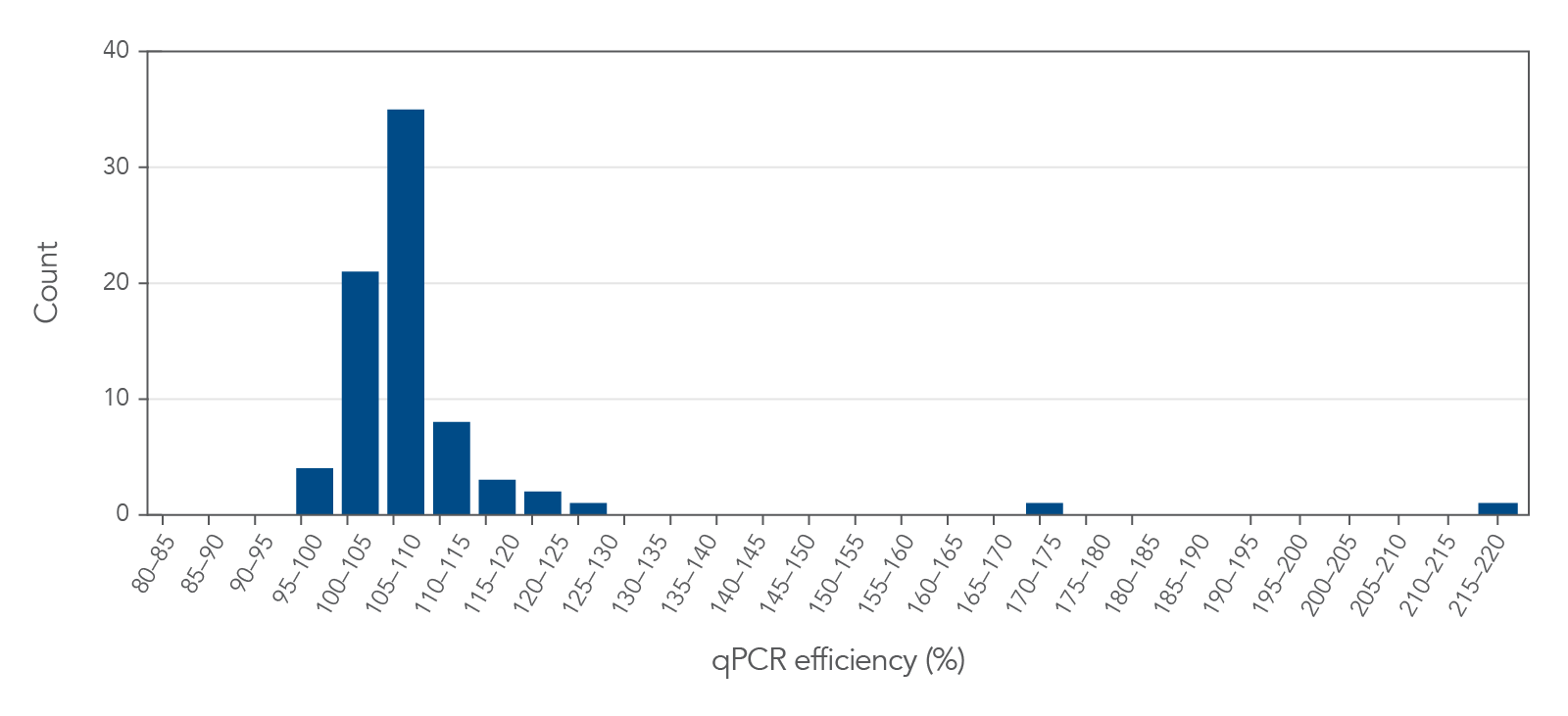
Figure 2. PrimeTime qPCR Primer Assays have an average reaction efficiency >90%. Sixty-one randomly selected PrimeTime qPCR Primer Assays and 15 PrimeTime qPCR Primer Assays for endogenous control genes used with SsoAdvanced Universal Probes Supermix (Bio-Rad) were analyzed over 5 sequential 4-fold dilutions (50–0.195 ng/reaction) of cDNA prepared from qPCR Human Reference Total RNA (Agilent). Reactions were run on the CFX384 Touch Real-Time PCR Detection System (Bio-Rad) using PCR cycling conditions: 30 sec 95°C; 40 x (15 sec 95°C, 30 sec 60°C). Average reaction efficiencies for the assays tested exceeded 98%.
Resources
Gene sets for common pathways
Frequently asked questions
How does the PrimeTime™ One-Step RT-qPCR Master Mix compare with one-step master mixes from other manufacturers?
How long can I leave my qPCR master mix (containing PrimeTime™ Gene Expression Master Mix) at room temperature and still generate consistent results?
The stability of PrimeTime Gene Expression Master Mix makes it ideal for overnight PCR runs and high throughput experiments using robotic liquid handling systems.
Reliable results have been obtained when the qPCR was run directly after setup or after the reactions were maintained at room temperature for up to 72 hr.
Download the white paper, Ambient shipping of PrimeTime Gene Expression Master Mix for more data.
Why does the reference dye come in a separate tube from the PrimeTime™ Gene Expression Master Mix?
We provide a separate tube of reference dye so that you may use the PrimeTime Gene Expression Master Mix on reference dye-dependent or dye-independent instruments.
Also, the separate tube of reference dye provides a way to add a high or low dye concentration to the master mix, depending on the requirements of your instrument.
Does the PrimeTime™ Gene Expression Master Mix use a hot-start polymerase?
Yes, it uses an antibody-mediated, hot-start polymerase. During the initial denaturation step of qPCR cycling, polymerase activity is unblocked when the polymerase is released from the antibody.
This hot-start polymerase enhances reaction precision of the qPCR by avoiding non-specific amplification of DNA and primer dimer formation. It is therefore critical to follow the cycling conditions provided in the PrimeTime Gene Expression Master Mix Protocol.
Is the Primetime™ Gene Expression Master Mix compatible with different real time PCR instruments or systems?
Reference dye is provided in a separate tube, so you may add it to the master mix at the levels required by your real-time PCR instrument.
How much reference dye should I add to the Primetime™ Gene Expression Master Mix?
The amount of reference dye that should be added to the PrimeTime Gene Expression Master Mix depends on which real-time PCR system you are using.
A table that details how much reference dye to add is provided in the PrimeTime Gene Expression Master Mix Protocol.
Does IDT offer VIC® probes for qPCR?
IDT offers the SUN™ dye, which is our product offering for the VIC dye (Thermo Fisher Scientific).
More information about probes labeled with the SUN fluorophore can be found on the PrimeTime™ qPCR probes page or the SUN fluorophore Decoded™ article.
How should I determine the annealing temperature for a probe-based qPCR assay? Should I select a melting temperature midway between my primer and probe melting temperatures (Tm)?
When performing qPCR it is ideal to have your probe Tm about 5−10 degrees higher than your primer Tm. The annealing temperature should be set 3−5 degrees lower than the lowest primer Tm.
Use this as a general guideline, but note that optimization may still be necessary.
What qPCR products does IDT ship at ambient temperature?
We ship PrimeTime™ Gene Expression Master Mix, PrimeTime qPCR assays, and other oligonucleotide products at ambient temperature.
Will phosphorothioate (PS) modifications on the ends of PCR primers provide nuclease resistance, and thus result in a longer half-life for the PCR product when transfected into a cell?
Phosphorothioate bonds (PS bonds) increase oligonucleotide resistance to exonuclease degradation. For single-stranded oligos, nuclease-resistance is highest when every phosphodiester bond is changed to a PS bond.
In general, we recommend at least 3 PS bonds on both 5' and 3' ends of the oligo, though more PS bonds will give greater exonuclease resistance.
Note that PCR primers with PS bonds at both ends will still result in double-stranded (ds) PCR products with PS bonds only at the 5' ends of each product strand. While protected from 5' to 3' nuclease activity, because these dsDNAs lack PS bonds at the 3' strand ends, exonucleases with 3' to 5' activity will still be able to degrade them.
Why do my primers have deleted 3' bases in the amplified fragment when I use a proofreading enzyme for PCR?
Proofreading polymerases (Pfu, Diamond Taq®, etc) have been shown to remove 3' bases from PCR primers [1].
If you experience this problem, you can protect your primer from degradation by adding one or more phosphorothioate bonds to the 3' end of your primer.
Also make sure to add the enzyme last, after the addition of dNTPs, in your PCR to minimize the risk of this issue taking place.
Reference
1. Skerra A. Phosphorothioate primers improve the amplification of DNA sequences by DNA polymerases with proofreading activity. Nucleic Acids Res. 1992;20(14):3551-3554.
Which dyes are compatible with my thermal cycler?
To determine what dyes are compatible with your instrument, download our Real-time qPCR assay design guide or take a look at the PrimeTime™ Multiplex Dye Selection Tool.
Which dyes are available for use with the IDT ZEN™ quencher?
Currently, the IDT ZEN quencher is available with FAM, HEX, TET, Yakima Yellow® (ELITech Group), JOE™ (Thermo Fisher Scientific), MAX™, and SUN™ fluorophores. Double-quenched probes have significantly lower background fluorescence, especially for longer probes.
The ZEN quencher is available on PrimeTime™ qPCR Probes and on the probes supplied with PrimeTime qPCR Probe Assays. For any special oligonucleotide requests that include the ZEN quencher but are not found on our website (non-catalog requests), please contact us.
For more information, see our DECODED™ article, Double-quenched probes increase qPCR sensitivity and precision.
Where on a nucleotide are Cy® dyes attached?
5′ Cy dyes are attached to the 5' hydroxyl group of the sugar via a phosphodiester bond.
3' Cy dyes are attached to the 3' hydroxyl group of the sugar via a phosphodiester bond.
Internal Cy dyes are attached to the backbone via the phosphodiester bonds of the bases between which the dyes occur.
When will my oligos be shipped?
IDT oligos have various estimated ship dates depending on product type and composition. Oligos added to the shopping cart will show an estimated ship date for each line item.
Once the order is placed, this estimated ship date can be found on the order confirmation email as well as in the order history of your web account. You can find the order history at idtdna.com/site/orderstatus/orderstatus.
Select the order number you wish to review, and the status and estimated ship date of each line item will be shown.
IDT will email you a confirmation when your order has been shipped. This shipping confirmation will include the tracking number of the package, which you can use to monitor the status of your shipment. If you have additional questions about your order, please contact us.
When preparing the qPCR reaction mix should I thaw or keep reagents (such as the Primetime™ Gene Expression Master Mix) on ice?
It is good practice, but not necessary to thaw or keep reagents on ice. We see no difference in results between reaction mixes prepared with the PrimeTime™ Gene Expression Master Mix that were used directly after thawing vs. those kept at room temperature for up to 24 hours.
PrimeTime Gene Expression Master Mix also shows exceptional temperature stability, even after a heat stress test (up to 8 hr at 55°C). Review the data in our white paper, Ambient shipping of PrimeTime Gene Expression Master Mix.
What type of purification should I choose for my qPCR primers?
Standard desalting is the recommended purification for qPCR primers.
What tools does IDT provide to design multiplex qPCR assays?
Use the free IDT OligoAnalyzer™ software to help ensure that primer and probe sets lack hairpins and homo- and heterodimer interactions. From there, you can link directly to the NCBI BLAST™ tool to further analyze possible sequence interactions. For help with using the BLAST tool, see Using BLAST to find primers—Tips from IDT.
For more information on setting up multiplex qPCR assays, see the article Recommended dye combinations for multiplex qPCR.
What is the structure of the ZEN™ internal quencher?
What is the stability of fluorescently labeled oligos and their fluorescent modifications?
The stability of fluorescently labeled oligos and fluorophore function is similar to the stability of a standard oligo.
IDT recommends oligos that are to be stored long term should be stored frozen at –20°C. If the oligos are going to be resuspended, storage in a TE buffer such as IDTE (10 mM Tris, 0.1 mM EDTA, pH 7.5–8.0) is better than using water (non-DEPC treated). The impact from the type of storage medium used is marginal at –20°C, but becomes more important at higher temperatures (see the DECODED™ article, Storing oligos: 7 things you should know, for the data).
With fluorophores, it’s important to remember that exposure to natural light or ozone can negatively impact stability. If fluorescently modified oligos will be exposed to sustained light, protective cover from the lighting source (e.g., using brown tubes and/or foil covering) should be considered.